
|
|
Research: Why? At 10,000 feet, our mission is to: improve the sustained use of land for natural resources, human well-being, and biodiversity. Management of soil biology is an untapped opportunity to accomplish this, so our research seeks to better understand how microbes function, particularly as they are subject to climate and land use change. While we focus on the biology of microorganisms, many of our projects are interdisciplinary and are complemented by outreach and education to tackle this mission.
Here are three themes of our lab's research. For a non-scientist’s description of the Evans Lab’s research, see Communication
Here are three themes of our lab's research. For a non-scientist’s description of the Evans Lab’s research, see Communication
1. How do microbes affect climate resilience of soils and crops?
 As land becomes more degraded and drought increases, microbes will alter their functioning and could buffer plants from drought.
As land becomes more degraded and drought increases, microbes will alter their functioning and could buffer plants from drought.
Like all organisms on earth, microbes will be subject to new climates, and we need to know how they will respond. We are particularly interested in microbial responses to changes in water availability. Many parts of the world will experience more drought and also more fluctuation in precipitation (more drought and flood). We are interested in how soil microbes will respond to these changes and the subsequent effect on a) soil biogeochemical cycles, like CO2 flux and b) plant growth under drought.
In the first theme (climate resilience of soil function): Microbially-mediated decomposition of plant litter and soil organic matter represents a major flux in the global carbon cycle, and microbial response to climate may determine whether soils are net emitters of CO2 or will sequester CO2, in the future. We are interested in how decomposing microorganisms behave in dry (including the extremely dry Namib desert) regions and how they respond to drought. Surprisingly, how heterotrophs respond to moisture is not well-described in ecosystem models, leading to potential errors as we simulate future carbon balance (see Evans et al. Functional Ecology).
Funding: NSF (DEB, Ecosystems).
In the second theme (climate resilience of crops): Drought and floods are a persistent concern to farmers, but will only increase in future climates. Microorganisms can increase water retention of soils, improving soil health, by producing more biofilms or sticky polysaccharides. They can also directly enhance water and nutrient uptake of plants, helping tolerate drought.
Funding: NSF (CNH2 interdisciplinary grant) and the KBS LTER, and USDA.
In the first theme (climate resilience of soil function): Microbially-mediated decomposition of plant litter and soil organic matter represents a major flux in the global carbon cycle, and microbial response to climate may determine whether soils are net emitters of CO2 or will sequester CO2, in the future. We are interested in how decomposing microorganisms behave in dry (including the extremely dry Namib desert) regions and how they respond to drought. Surprisingly, how heterotrophs respond to moisture is not well-described in ecosystem models, leading to potential errors as we simulate future carbon balance (see Evans et al. Functional Ecology).
Funding: NSF (DEB, Ecosystems).
In the second theme (climate resilience of crops): Drought and floods are a persistent concern to farmers, but will only increase in future climates. Microorganisms can increase water retention of soils, improving soil health, by producing more biofilms or sticky polysaccharides. They can also directly enhance water and nutrient uptake of plants, helping tolerate drought.
Funding: NSF (CNH2 interdisciplinary grant) and the KBS LTER, and USDA.
|
Methods highlight: Field climate manipulations: We examine effects of drought experimentally by inducing drought with rainout shelters. We have a large project examining rainfall exclusion in collaboration with 9 (!) other labs, at Kellogg Biological Station LTER (REX). We also leverage DroughtNet, a coordinated experiment of rainout shelters, in an NSF-funded project to examine microbial response to drought at 35 sites across the world. We study both managed and natural ecosystems.
|
2. What ecological processes control microbial composition and function in agricultural lands?
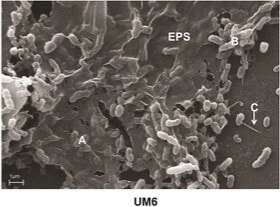 Pure culture of bacteria embedded in biofilm "EPS" matrix. Biofilms may increase soil structure, carbon, and water holding capacity.
Pure culture of bacteria embedded in biofilm "EPS" matrix. Biofilms may increase soil structure, carbon, and water holding capacity.
Soil microbial communities are made up of tens of thousands of species across multiple domains of life, interacting on a micro-scale. Processes that control the composition and function of communities, and their response to agricultural practices, needs further study. Two processes we focus on are (a) microbial dispersal at landscape scales (b) micro-scale function
In the first theme (microbial dispersal at landscape scales): Recently, dispersal has received greater recognition as a process shaping microbial community composition. However, the specifics are still unclear. How much does dispersal matter, compared to other things like species filtering? What microbial species are dispersal limited and which are not? We study dispersal in agricultural systems, since changes in land use can alter the sources and sinks of microbial communities, and have showed dispersal maintains diversity in these systems (Evans et al. 2016, ISMEJ, and Evans et al. 2020, Env Micro). Specifically, we are interested in prairie strips (strips of native vegetation that are introduced into row crops) which may introduce beneficial microorganisms into agricultural lands via dispersal.
In the second theme (micro-scale function):, we ask how micro-scale dynamics influence microbial function. For instance, how colonizing microorganisms, specifically functions coded on their genetic material, move through extant communities through replication and horizontal gene transfer. We are also interested in microbial production of biofilms since this can affect organic matter accumulation, microbial interactions, and drought tolerance.
Funding: KBS Long Term Ecological Research Station (LTER), and the Great Lakes Bioenergy Research Center (GLBRC).
In the first theme (microbial dispersal at landscape scales): Recently, dispersal has received greater recognition as a process shaping microbial community composition. However, the specifics are still unclear. How much does dispersal matter, compared to other things like species filtering? What microbial species are dispersal limited and which are not? We study dispersal in agricultural systems, since changes in land use can alter the sources and sinks of microbial communities, and have showed dispersal maintains diversity in these systems (Evans et al. 2016, ISMEJ, and Evans et al. 2020, Env Micro). Specifically, we are interested in prairie strips (strips of native vegetation that are introduced into row crops) which may introduce beneficial microorganisms into agricultural lands via dispersal.
In the second theme (micro-scale function):, we ask how micro-scale dynamics influence microbial function. For instance, how colonizing microorganisms, specifically functions coded on their genetic material, move through extant communities through replication and horizontal gene transfer. We are also interested in microbial production of biofilms since this can affect organic matter accumulation, microbial interactions, and drought tolerance.
Funding: KBS Long Term Ecological Research Station (LTER), and the Great Lakes Bioenergy Research Center (GLBRC).
|
|
Methods highlight: individual-based models. We have used models to study both microbial assembly and microbial linkages to ecosystem function. The movie to the left shows a simulation of microbial cells on a soil micro-grid (1 square milimeter!). Different colors represent different microbial functional groups (e.g. cheaters). Individuals compete and are filtered on the grid over time. We used this model to study the role of microbial physiology - and community composition - in drying-rewetting pulses (Evans et al. 2015) with Tina Kaiser at the International Institute of Applied Systems Analysis.
|
3. What microbial processes influence plant nitrogen availability? Can we bolster them in degraded croplands?
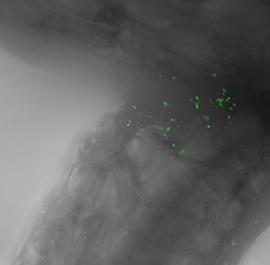 The nitrogen (N) fixing bacteria Azospirillum brasilense (green dots), which tend to colonize the space at the intersection of a switchgrass root.
The nitrogen (N) fixing bacteria Azospirillum brasilense (green dots), which tend to colonize the space at the intersection of a switchgrass root.
Nitrogen is a major limitation to both natural and managed lands. While we alleviate this limitation with fertilizer, production and application causes negative environmental consequences. It is particularly undesirable in bioenergy system because (i) high fertilizer application can negate any potential climate mitigation that bioenergy growth might contribute and (ii) bioenergy may be most promising to grow on marginal, degraded lands where it does not compete with food. We are examining how the rhizosphere microbiome can provide nitrogen to perennial grasses that can be used for cellulosic bioenergy production. Plants such as switchgrass could stimulate beneficial N-fixing organisms by secreting exudates from their roots, but may have less incentive to do this when fertilized, decreasing the N-fixing capacity of the soil.
Funding: DOE, both the Great Lakes Bioenergy Research Center (GLBRC) and the BER.
See press on this grant in MSU Today
See pictures from the field on our Field Pictures page.
Funding: DOE, both the Great Lakes Bioenergy Research Center (GLBRC) and the BER.
See press on this grant in MSU Today
See pictures from the field on our Field Pictures page.
|
Methods highlight: stable isotopes. Stable isotopes are a powerful way to link microbial composition and function and to study and trace plant-microbe interactions. We use 15N to understand how nitrogen gas is fixed into microbial biomass and plant biomass. We also use 13CO2 labeling to quantify rates of root exudation and stable isotope probing (SIP) to characterize which microbes use which C and N compounds.
|
See Publications for Evans citations


